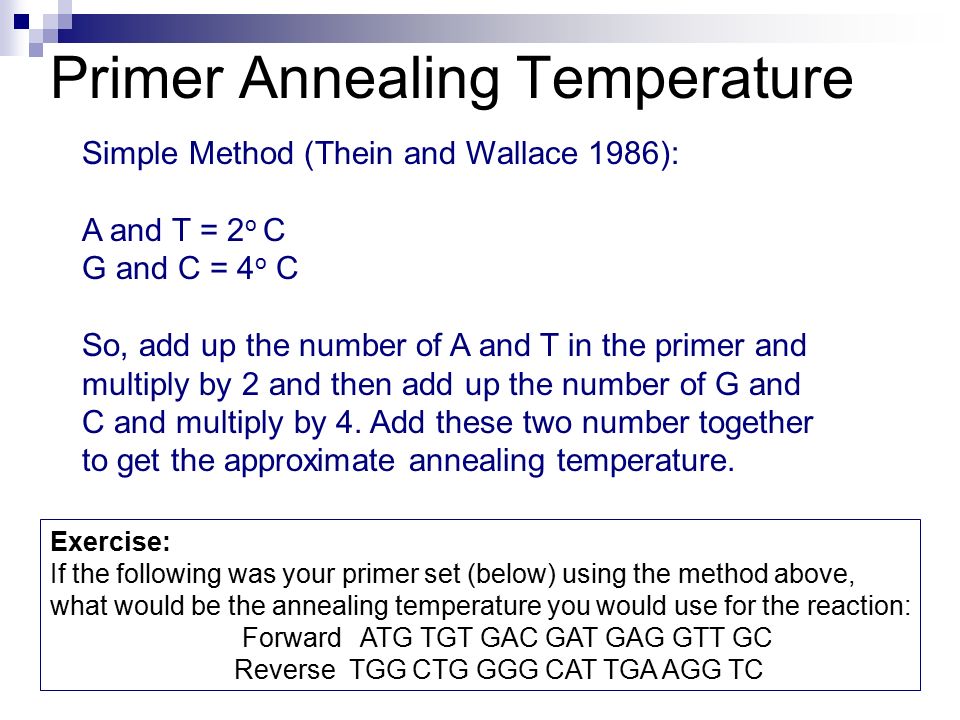
How To Calculate Annealing Temperature. Compare observed annealing temperature to calculated annealing temperature. Two standard approximation calculations are used. Imental protocol to test actual annealing temperature is described visualize the PCR products in a horizontal agarose gel electrophoresis 5. χ T t S π min t 1 N min t exp δ t T t S π min t 1 N min t t S exp E max t T t S exp E min t T.

Lets take a look at an example of how to calculate. Tm melting temperature in C A number of adenosine nucleotides in the sequence T number of thymidine nucleotides in the sequence C number of cytidine nucleotides in the sequence. Tm wAxT 2 yGzC 4 where wxyz are the number of. Where T m of primer is the melting temperature of the less stable primer-template pair and T m of product is the melting temperature of the PCR product 1. The annealing temperature Ta chosen for PCR relies directly on length and composition of the primers. Extension temperature recommendations range from 6575C and are specific to each PCR polymerase.
Gradient PCR was used in the next experiment in order to determine the optimal annealing temperature.
Primer length is typically between 18 and 22 nucleotides. Extension rates are specific to each PCR polymerase. Imental protocol to test actual annealing temperature is described visualize the PCR products in a horizontal agarose gel electrophoresis 5. χ T t S π min t 1 N min t exp δ t T t S π min t 1 N min t t S exp E max t T t S exp E min t T. Determine the quantity and size of the PCR product for each set of primers and 6. Tm product is the melting point of the product.