
E Crisp Fast Crispr Target Site Identification. In CRISPR-SE all gRNAs are virtually compared with query gRNA. The design process incorporates different parameters of how CRISPR. CRISPRdirect predicts specific gRNAs based on in silico prediction of specificity but the lack of implementation of evidence-based metrics to predict off-target sites is a notable caveat. The task of E-CRISP is to find CRISPR target sites which are specific.
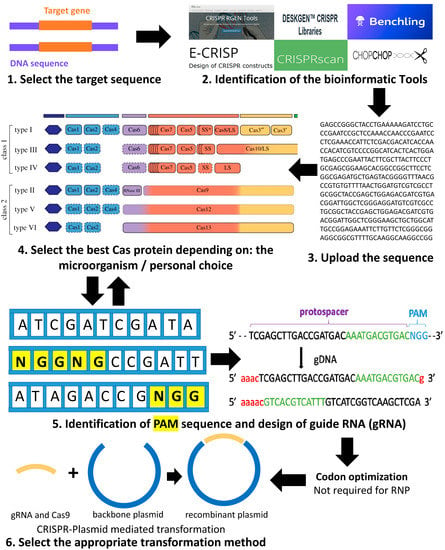
AU - Boutros Michael. The design process incorporates different parameters of how CRISPR. CRISPR Clustered Regularly Interspaced Short Palindromic Repeats-based gene editing has been widely implemented in various cell types and organisms. E-CRISP is a computational tool to design and evaluate guide RNAs for use with CRISPRCas9. Fast CRISPR target site identification Florian Heigwer Grainne Kerr and Michael Boutros Here we describe E-CRISP a web application to design gRNA sequences. Our comprehensive benchmark study of these existing resources provides insightful guidance for off-target effect research in four aspects.
Here we present E-CRISP a.
AU - Boutros Michael. E-CRISP can also be used to reevaluate CRISPR constructs for onor off-target sites and targeted genomic loci. If it can map the gRNA to another. Several tools are available for sgRNA design while limited tools were. Is supported by a European Research Council Advanced Grant of the European. We found at least one gRNA design per locus.